| Pairwise | MSAdb | MSA... |
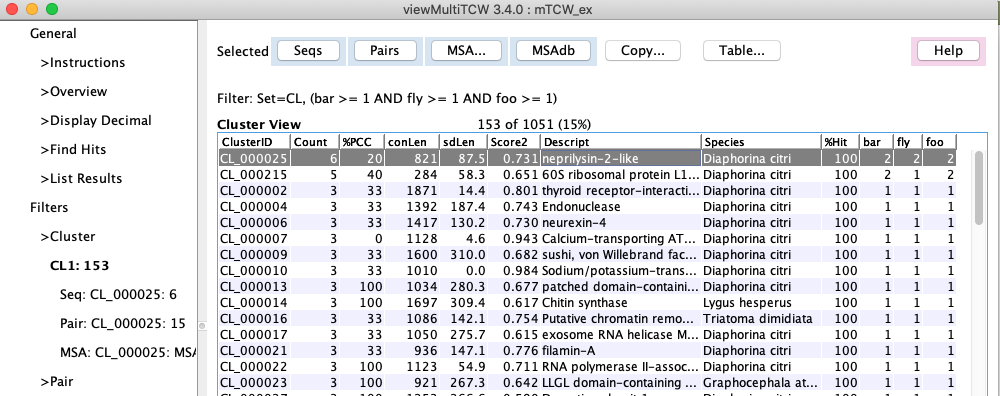
Cluster table with MSA

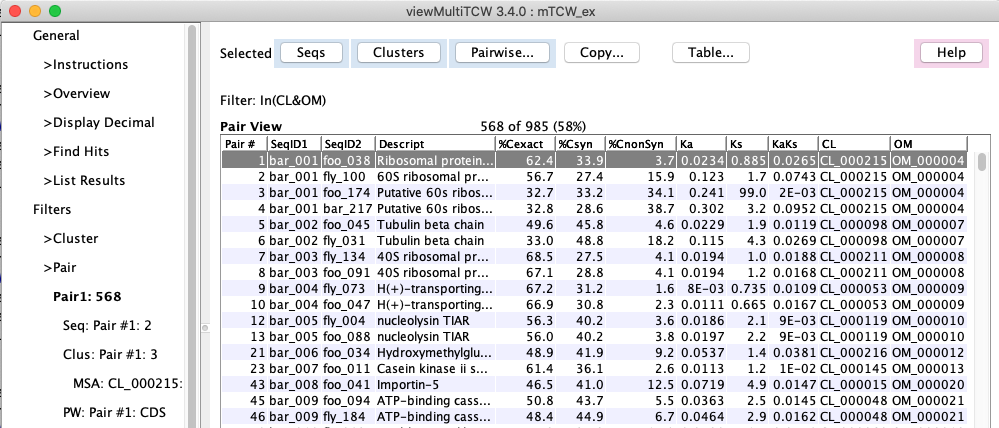
| Pair table with Pairwise

|
If there are only protein (AA) sequences in the database, only the AA options described below will be available.
Pairwise... | Go to top |
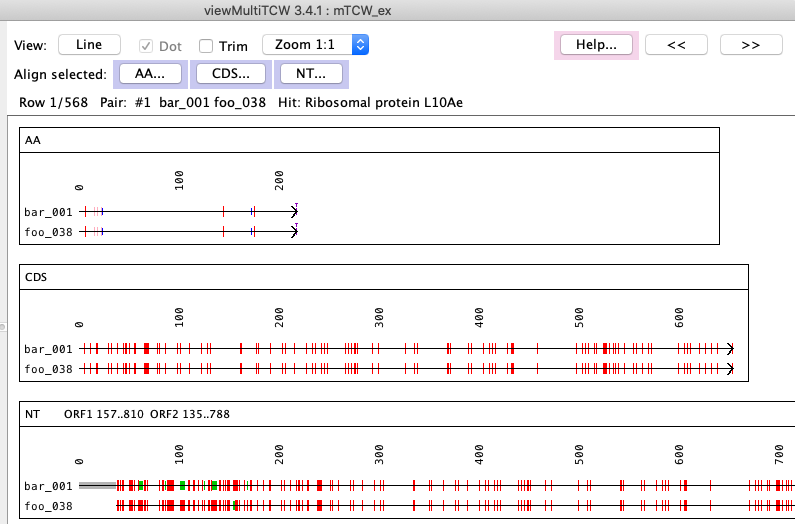
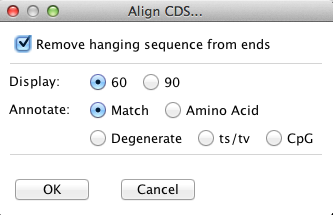
Menu for the CDS text alignment:
|
Text alignment of the CDS showing matches.
The '|' are mutations in synonymous codons, the '*' mutations in nonsynonymous codons. 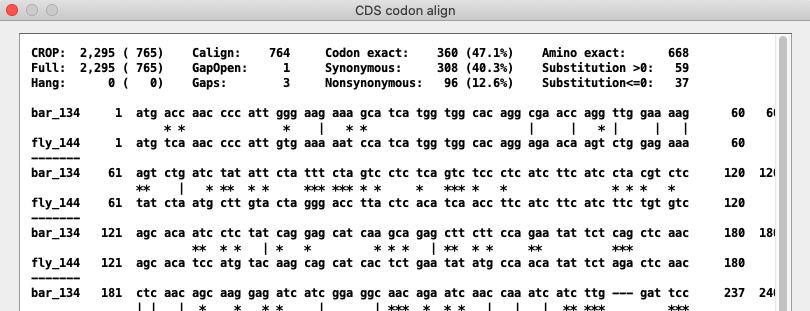
|
MSAdb | Go to top |
MSA... | Go to top |
| The AA is the translated ORF, CDS is the un-translated ORF, and NT is the entire transcript. | 
|
| Go to top |