|
||

|
SyMAP - Synteny Mapping and Analysis Program | |
|
||
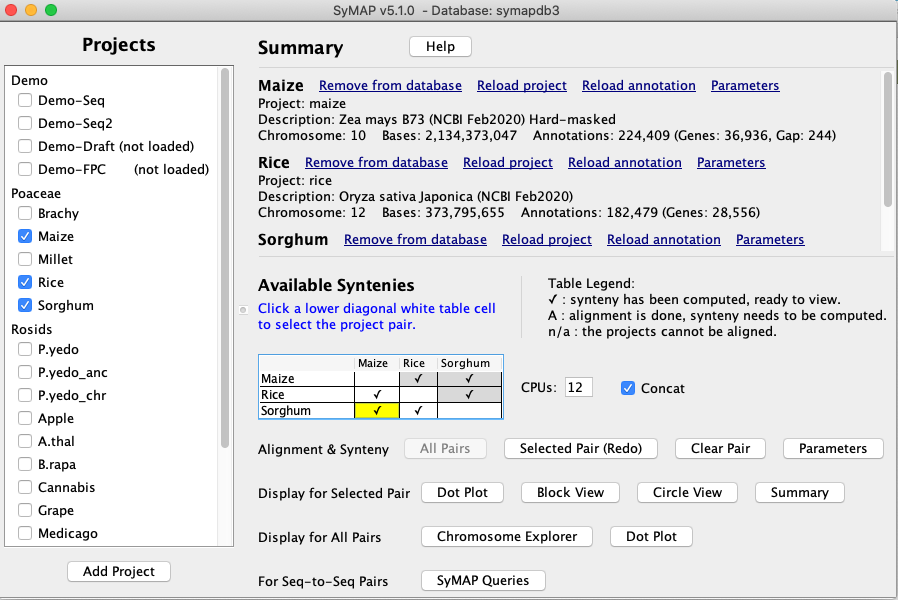
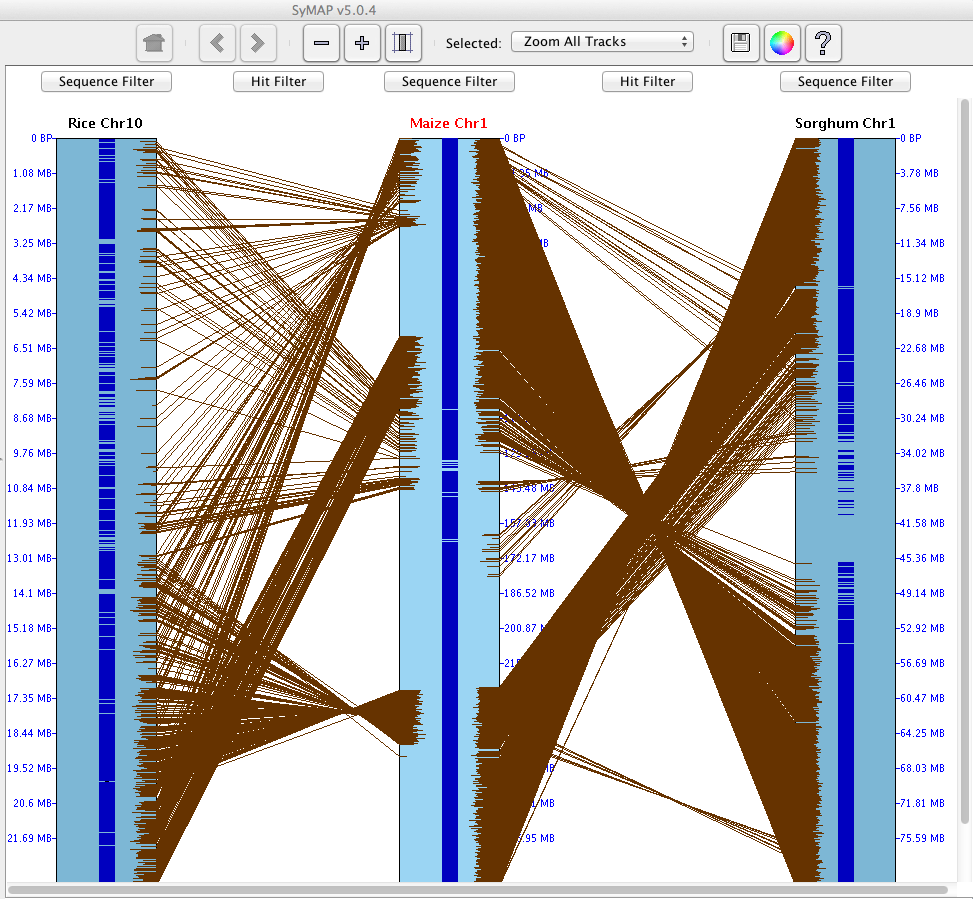
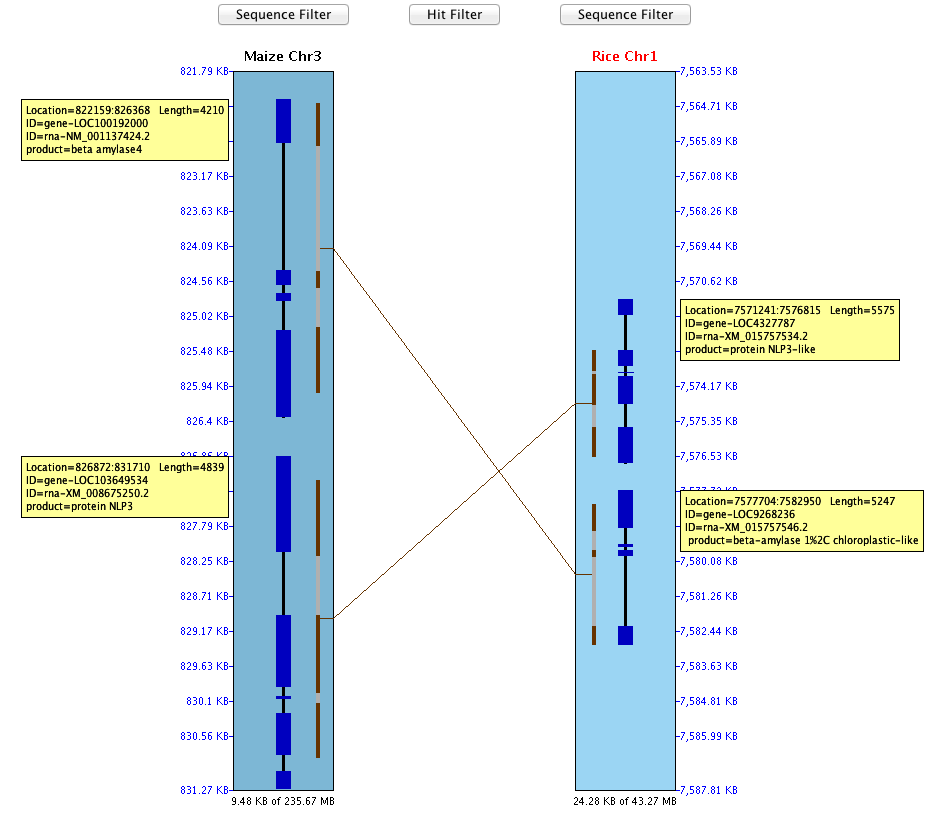
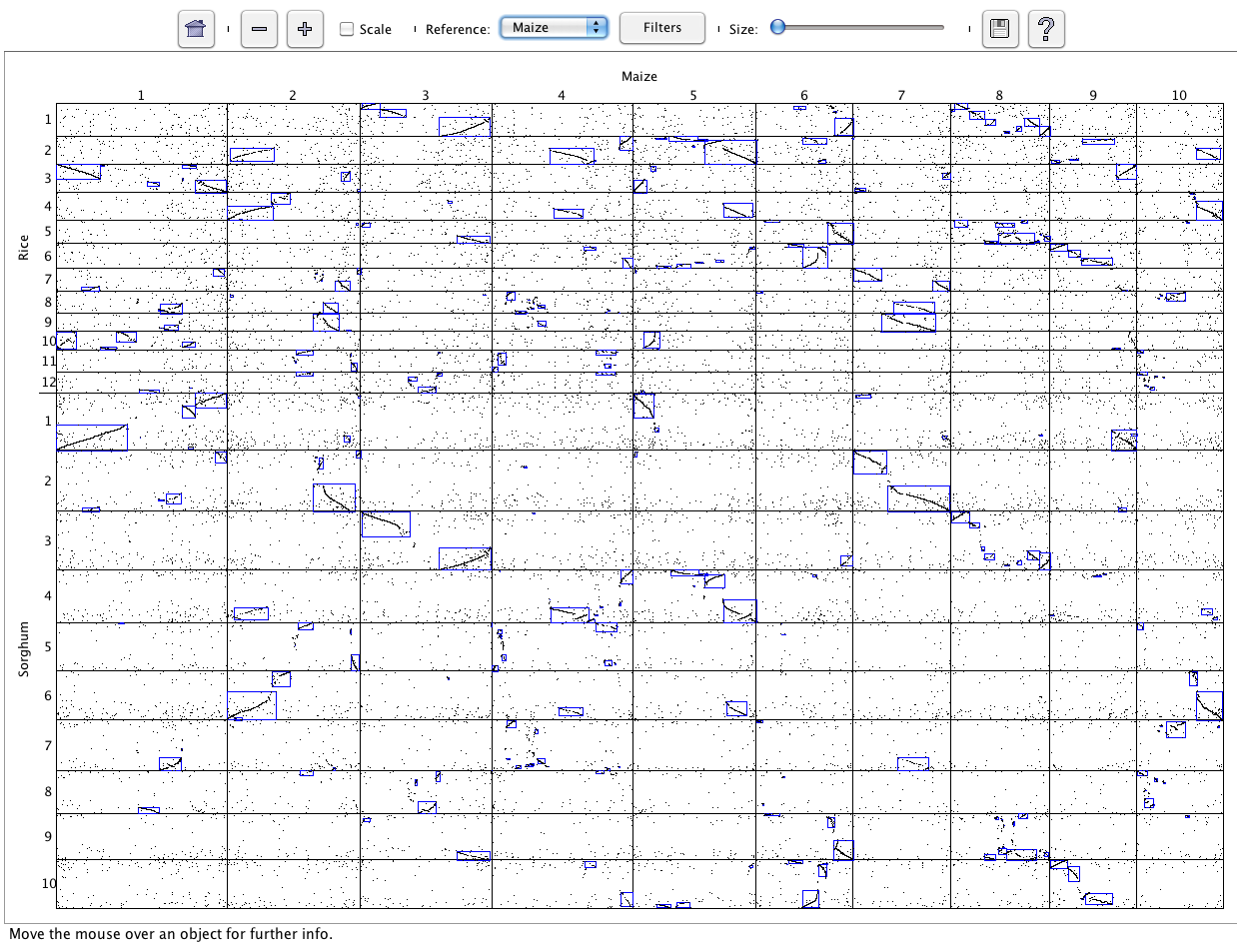
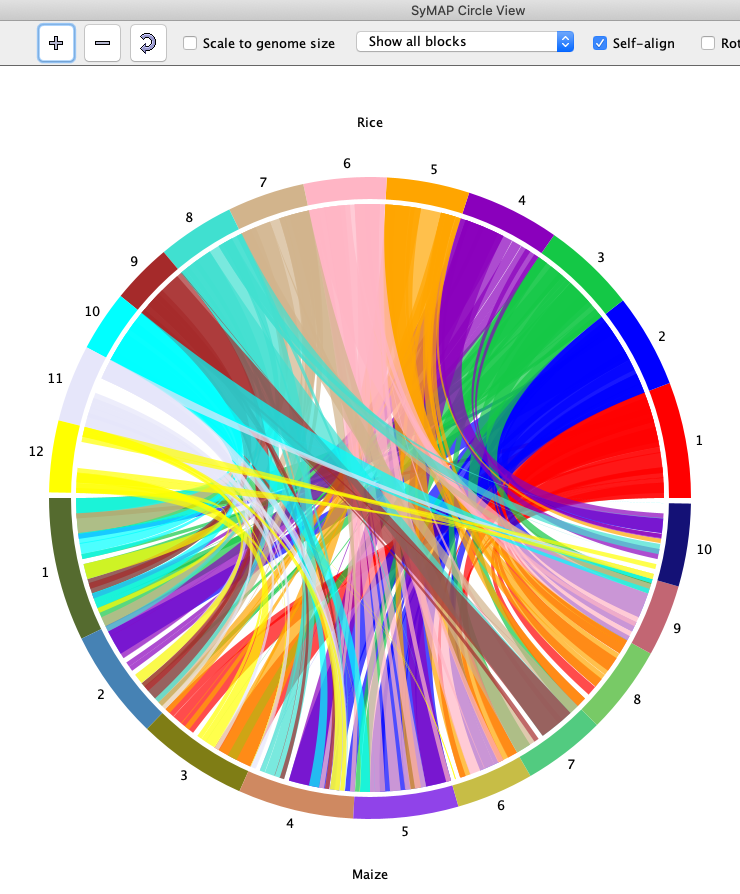
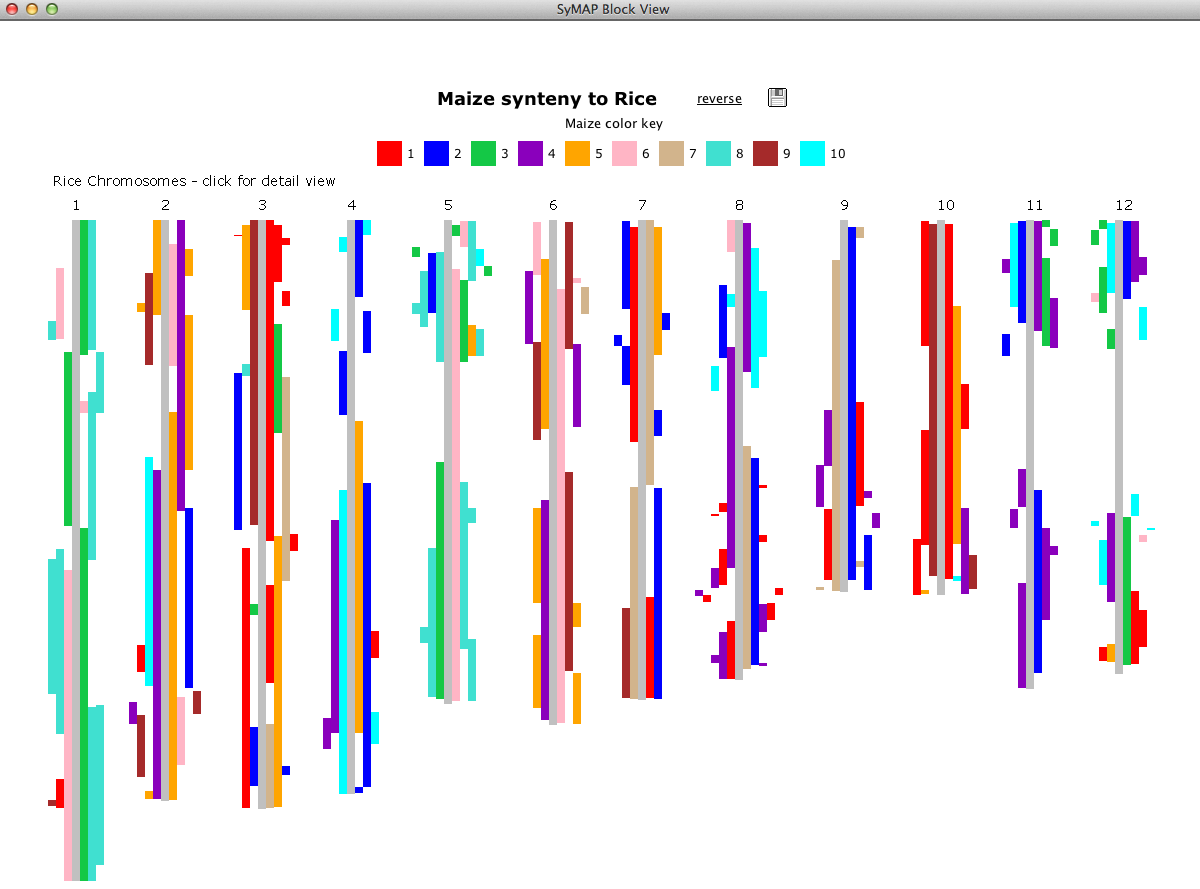
SyMAP (Synteny Mapping and Analysis Program) is a software package for
detecting, displaying, and querying syntenic relationships between sequenced genomes.
 It is designed for medium-to-high divergent eukaryotic genomes (not bacteria).
It is designed for medium-to-high divergent eukaryotic genomes (not bacteria).
 It can align a draft genome to a fully sequenced genome
(see peach), but not draft-to-draft.
It can align a draft genome to a fully sequenced genome
(see peach), but not draft-to-draft.
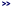 It can perform self-synteny. It can align to FPC physical maps.
It can perform self-synteny. It can align to FPC physical maps.
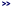 It is designed for medium-to-high divergent eukaryotic genomes (not bacteria).
It is designed for medium-to-high divergent eukaryotic genomes (not bacteria).
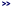 It can align a draft genome to a fully sequenced genome
(see peach), but not draft-to-draft.
It can align a draft genome to a fully sequenced genome
(see peach), but not draft-to-draft.
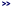 It can perform self-synteny. It can align to FPC physical maps.
It can perform self-synteny. It can align to FPC physical maps.
 Releases v5.1.0 and later are available from the Github links below.
Releases v5.1.0 and later are available from the Github links below.
|
|
||||||||||||||||||||||||||||||
 |
This project was partially supported by the National Research Initiative of USDA's National Institute of Food and Agriculture, grant #0214643. |
| All upgrades starting with release 7-Nov-2016 have been developed by CAS without funding. | |
References
C. Soderlund, M. Bomhoff, and W. Nelson (2011) SyMAP v3.4: a turnkey synteny system with application to plant genomes Nucleic Acids Research 39(10):e68 Link
C. Soderlund, W. Nelson, A. Shoemaker and A. Paterson (2006) SyMAP: A system for discovering and viewing syntenic regions of FPC maps. Genome Research 16:1159-1168. Link